Warning
This is still a draft! Please refer to the PDF version.
4. Dockerizing DraCor, for Example. On Versioning Programmable Corpora#
I make no claims to elegance in programming, but I am confident that the scripts work, at least as of today.
—Andrew Piper, Foreword to Enumerations
4.1 Containerizing a Research Environment#
As already noted in the introduction (see 2. Introduction: The Problem of Reproducibility in Dynamic Digital Ecosystems), the use of a version control system like Git can be seen as both a powerful and user-friendly mechanism to stabilize “living” corpora. However, “programmable” corpora consist of a connection of data (“living corpus”) with a lightweight research software (in the form of a research-driven API) whereby this connection can be conceptualized as a distributed research infrastructure in a dynamic digital ecosystem (cf. Börner and Trilcke [2023]). Such a research infrastructure can no longer be stabilized only via the versioning mechanism of Git, since in addition to the two components (data, code), their specific connection is also crucial for the possibility of reproducing the research conducted with it. For such scenarios, as will be shown below, a container-based approach is suitable, whereby we will rely on containerization using the Docker software.
Our approach is inspired by the concept of research artifacts from computer science, which is described by Arvan et al. as “self-contained packages that contain everything needed to reproduce results reported in a paper” and which are also “typically self-executable, meaning that they are packaged within a virtual machine […] or within a container” [Arvan et al., 2022]. The prototypical implementation, on which we will report in the following, is again based on the Drama Corpora Platform, DraCor. In a DraCor-based experiment, we set ourselves the challenge of making a network-analytic study, which we conducted for a paper publication, as fully replicable as possible using Docker.
The containerization technology “Docker” is widely used in the IT industry, because it can speed up development cycles and can reduce overhead when deploying applications. Especially in the field of “DevOps” – “Dev” for development and “Ops” for operations, which refers to the processes that are necessary to have an application run on a server – the importance of communication between the development and the operations team is considered highly important. Thus Docker workflows have been introduced. They do not only streamline communication processes, they also shift the responsibility of handling software dependencies to the development team: Docker enables the people actually writing the application to specify the environment in which their software should be run. There are a couple of key components to such workflows: The so-called “Dockerfile”[1] is a meaningful and executable form of documentation. It contains the steps necessary to build a highly portable, self-contained digital artifact, a “Docker image”[2]. These images can be easily deployed on a designated infrastructure as “Docker containers”[3].
Certainly, in CLS research projects we will rarely find teams of development and operation professionals that are in need of communicating better. But, we would like to argue that still the attempt of reproducing research could be framed in a similar sense: On the one hand we have an individual researcher (or a team of researchers) that conducts a study. In our analogy, these are the developers. Their product – a study – relies on some application or script that operates on data. On the other hand we have researchers wanting to reproduce or verify the results, similar to operations professionals that have to deploy someone else’s application on a server.
It becomes evident that some hurdles in the process of reproducing CLS research exist due to a lack of clear communication on how to run the analysis scripts and a tendency to offload the responsibility of setting up an environment in which the analysis could be executed to the reproducing party. A containerized research environment might circumvent these problems: Instead of claiming that scripts “work, at least as of today” on the machine of the developing researcher, as for example, Andrew Piper writes it in his foreword to his book “Enumerations” [Piper, 2018, xii]; when employing container technology, it could be guaranteed instead that a container created from an image containing a runnable self-contained research environment can be re-run. This would allow for a reproduction of the study. Instead of saying: The analysis was run-able on my machine when writing the paper and publishing the code only, in addition a researcher could provide a run-able research artifact alongside the study. When using Docker, this could be one or more images that would allow to re-create the research environment the study was conducted in.[4]
In the following, we will report on our exemplary experiment in making a CLS study replicable by using Docker technology.[5]
4.2 Case Study: Dockerizing a Complete CLS Study#
We exemplify the benefits of a Docker-based research workflow by referring to our study “Detecting Small Worlds in a Corpus of Thousands of Theater Plays” [Trilcke et al., 2024]. In this study, we tested different operationalizations of the so-called “Small World” concept based on a multilingual “Very Big Drama Corpus” (VeBiDraCor) of almost 3,000 theater plays. As explained above, the corpora available on DraCor are “living corpora” – which means that both the number of text files contained and the information contained in the text files changes (e.g. with regard to metadata or mark-up). This poses an additional challenge for reproducing our study. Furthermore, our analysis script (written in R) retrieves metadata and network metrics from the REST API of the “programmable corpus”. Thus, we had to devise a way of not only stabilizing the corpus but also the API.
DraCor provides Docker images for its services, which are, at its core the API, a frontend, a metrics service, that does the calculation of the network metrics of co-presence networks of the plays, and a triple store. The ready-made Docker images can be used to set up a local DraCor environment.[6]
For VeBiDraCor we devised a workflow that spins up a Docker container from a versioned bare Docker image of the DraCor database[7] and ingests the data of the plays downloaded (“pulled”) from specified GitHub commits using a Python script[8]. We then committed this container with docker commit[9] to create a ready to use Docker image of the populated database and API (see Fig. 8).
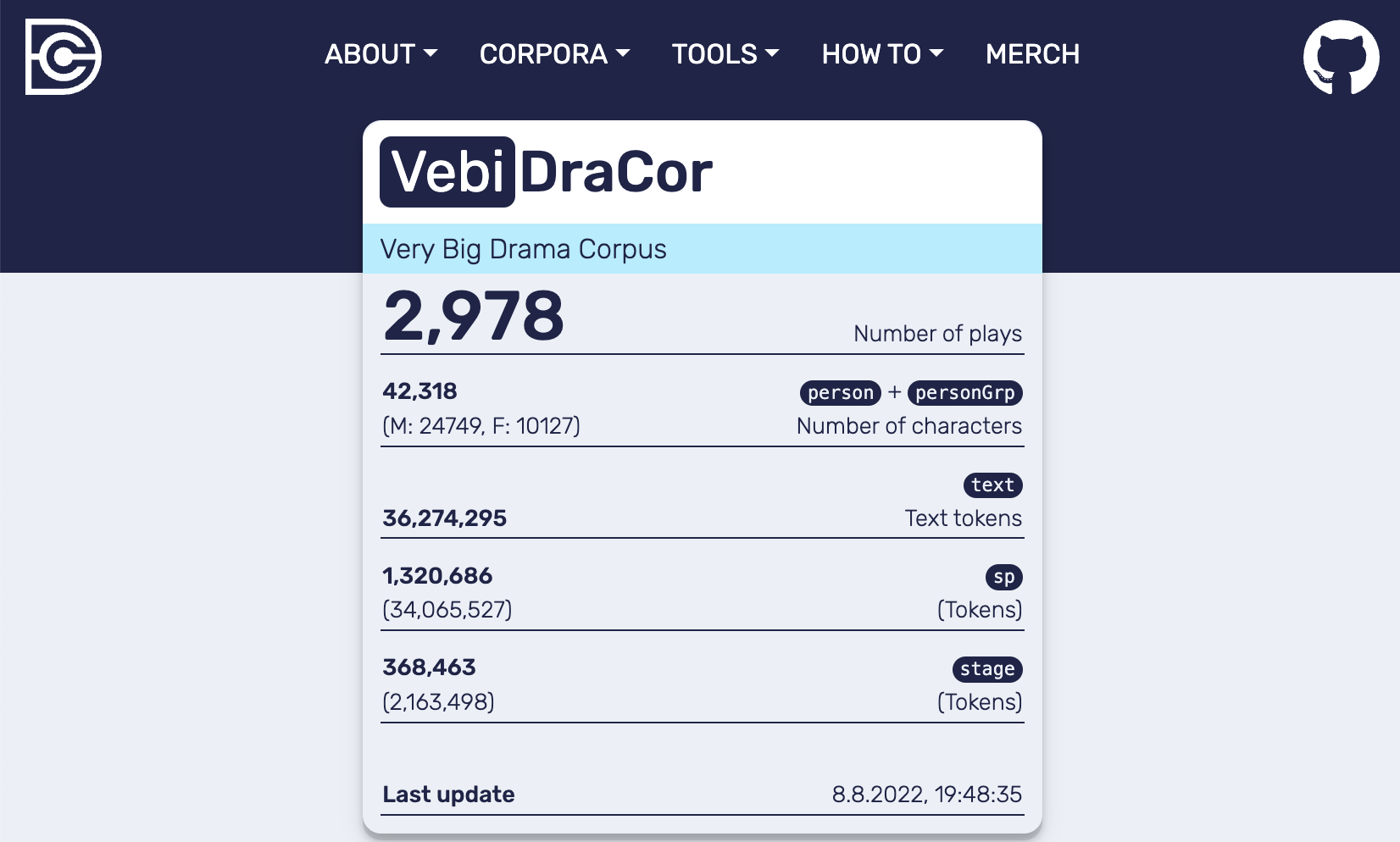
Fig. 8 Frontend of the local DraCor infrastructrue with added corpus VeBiDraCor#
Because the build process is modular and documented in a Dockerfile, it is also possible to quickly change the API’s base image or the composition of the corpus by editing a manifest file that controls which plays from which repositories at which state are included.
In a second step, we also dockerized the research environment: a Docker container running RStudio to which we added our analysis script. The preparation of this image is documented in a Dockerfile. As base image we used an image of the rocker-project. We used docker commit to “freeze” this state of our system and published all images. We call this state the “pre-analysis state”, which is documented in a Docker Compose file.
After we ran the analysis, we again created an image of the RStudio container with docker commit, thus turning it into a Docker image in which basically “froze” the state of the research environment after the R-script was run. The image of this “post-analysis state” was also published on the DockerHub repository. It allows for inspection and verification of the results of our study in the same environment that we used (see Fig. 9).[10]
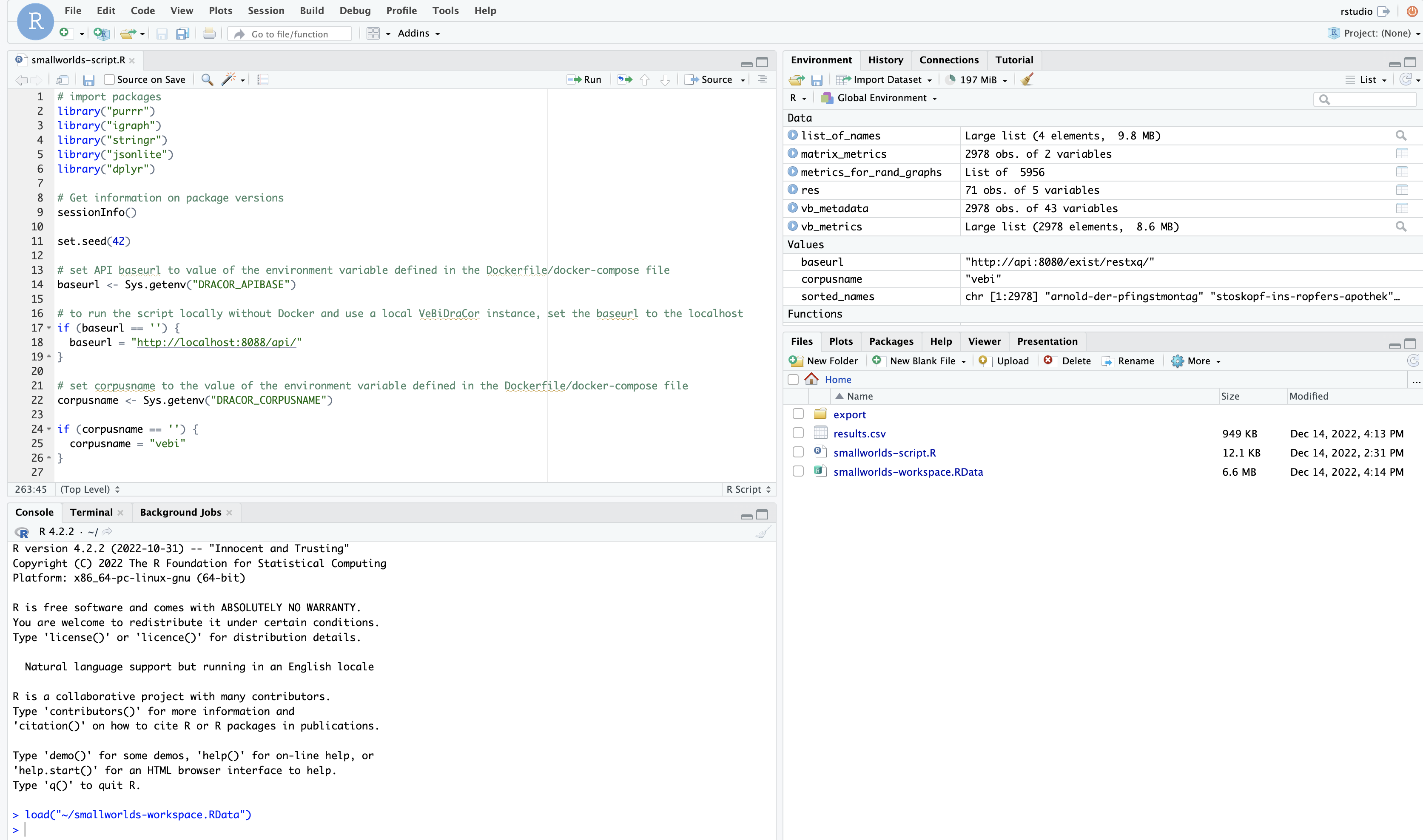
Fig. 9 RStudio after the analysis was run#
By having these images representing two moments in the course of our analysis, we not only make our analysis transparent, we also allow for different scenarios of repeating our research. For example, starting an environment with the Docker Compose file that documents the “pre-analysis state” would allow a researcher to exactly repeat our analysis by re-running the script on the exact same data.
But also other scenarios of repeating research (e.g. “replication”, “reproduction”, “revision”, “reanalysis”, “reinvestigation”; cf. [Schöch, 2023]) could be implemented easily. To give one example: A researcher could adapt the Jupyter Notebook we used to assemble “VeBiDraCor” and create an image of the local corpus container the same way we did. By changing a single line in the Docker Compose file documenting the “pre-analysis state” it is possible to start the whole system with this different data. For running our R-script to analyze this data he or she could still use a container created from our RStudio image and thus run the analysis the exact same way we did, but on different data.
The following code cell demonstrates how to re-create the intrastructure that was used to generate the results reported in the paper.[11]
%%script bash --bg
# Clone the GitHub repository containing the data of the study
git clone https://github.com/dracor-org/small-world-paper.git
# Go into the just downloaded repository and switch to the branch "publication-version"
cd small-world-paper
git checkout publication-version
# Start the infrastructure in the "post-analysis-state" as defined
# in the Compose file "docker-compose.post.yml"
docker compose -f docker-compose.post.yml up
Show code cell content
%%bash
# To see the status of the containers
# the STATUS of the container derived from the image ingoboerner/vebidracor-api:3.0.0 should be "Up"
docker ps -a
CONTAINER ID IMAGE COMMAND CREATED STATUS PORTS NAMES
c8be87a4cfb1 ingoboerner/dracor-frontend:v1.4.3_local "/docker-entrypoint.…" Less than a second ago Created small-world-paper-frontend-1
74ec2b952ffd ingoboerner/smallworld-rstudio:dcac262 "/init" Less than a second ago Created small-world-paper-rstudio-1
633f3176788e ingoboerner/vebidracor-api:3.0.0 "java org.exist.star…" Less than a second ago Created small-world-paper-api-1
9c91409e298c dracor/dracor-frontend:v1.6.0-dirty "/docker-entrypoint.…" 4 minutes ago Up 4 minutes 0.0.0.0:8088->80/tcp chiasmus_detection-frontend-1
c9cef3212e75 dracor/dracor-api:v0.90.1-local "./entrypoint.sh" 4 minutes ago Up 4 minutes (healthy) 0.0.0.0:8080->8080/tcp chiasmus_detection-api-1
f2e4c6da0e08 dracor/dracor-metrics:v1.2.0 "pipenv run hug -f m…" 4 minutes ago Up 4 minutes 0.0.0.0:8030->8030/tcp chiasmus_detection-metrics-1
307487cc4a2c dracor/dracor-fuseki:v1.0.0 "/usr/bin/tini -- /e…" 4 minutes ago Up 4 minutes 0.0.0.0:3030->3030/tcp chiasmus_detection-fuseki-1
4.3 Simplifying the Workflow: StableDraCor#
The workflow presented in the section 4.2 Case Study: Dockerizing a Complete CLS Study above is still quite complex. There are multiple steps involved to set-up the locally running infrastructure some of which need to be run from the command line. In addition, one has to create a bare container with a database and an API, populate it with the data, conduct the analysis, create several Docker images, and, ultimately, publish them in a repository, e.g. on DockerHub. There is also the need to create the Docker Compose file that specifies which system components are needed to recreate the environment the analysis was run in at different points in time. Thus, there is a considerable need to make the process more user-friendly.
Our approach to simplifying the process focuses on developing a Python package called “StableDraCor” that makes the setting-up of local DraCor instances and populating them with data easier by somewhat “hiding” the complexity of the Docker and Docker Compose commands. While there is no real need for a generic tool managing containers and images (because this can be done with Docker Desktop), with “StableDraCor” we address the complexity of setting up the specific DraCor infrastructure components and loading DraCor corpora (or a subset thereof).
In the following, we describe the workflow using the tool. The package can be built and installed from the GitHub repository[12].
Show code cell content
%%bash
# Clone the repository
# The target directory is outside of the current working directory "report"
git clone https://github.com/ingoboerner/stable-dracor.git /home/d73/stable-dracor
# Switch into repository and checkout a commit
# We checkout the commit because the package is still in development and we want to install a
# certain version to be used here
# the -c option is optional; it suppresses a warning about git being in so-called detached head state
cd /home/d73/stable-dracor
git -c advice.detachedHead=false checkout df31c4e
# install the package
pip install .
# cleanup: remove the previously cloned repository
cd /home/d73/report
rm -rf /home/d73/stable-dracor
After initializing a StableDraCor instance, the infrastructure can be started with a single command run(). If a user does not specify any parameters (like pointing to a designated custom local Docker Compose file) the script fetches a configuration of the system specified by a Docker Compose file (compose.fullstack.yml) and starts the defined containers.
Show code cell content
# Import the package
from stabledracor.client import StableDraCor
# Use a GitHub Personal Access Token, see
# https://github.com/ingoboerner/stable-dracor/blob/e300d77c419537538b4d491a8bbe2b9449123131/notebooks/03_faq.ipynb
import os
github_token = os.environ.get("GITHUB_TOKEN")
# Initialize a local DraCor infrastructure
# Provide metadata like a "name" and a description
local_dracor = StableDraCor(
name="my_local_dracor",
description="My local demo DraCor system",
github_access_token=github_token
)
# Start the infrastructure
local_dracor.run()
After running the above cell in the “Executeable Report” the local DraCor frontend becomes available at http://localhost:8088. As can be seen in Frontend of the local DraCor infrastructure with no corpora loaded no corpora have been loaded yet.

Fig. 10 Frontend of the local DraCor infrastructure with no corpora loaded#
The package supports setting-up local custom corpora either by copying a corpus or parts thereof from any running DraCor system, for example the production system or the staging server, containing even more corpora that are currently prepared for publication.
In the following code cell the “Tatar Drama Corpus” from the production instance of DraCor to the local database (see Frontend of the local DraCor infrastructure with no corpora loaded).
Show code cell content
local_dracor.copy_corpus(source_corpusname="tat")
After executing the command in the code cell above there will be a single corpus in the local DraCor instance (see Frontend of the local DraCor infrastructure with the “Tatar Drama Corpus” loaded) that is a copy of the data currently available at https://dracor.org/tat.
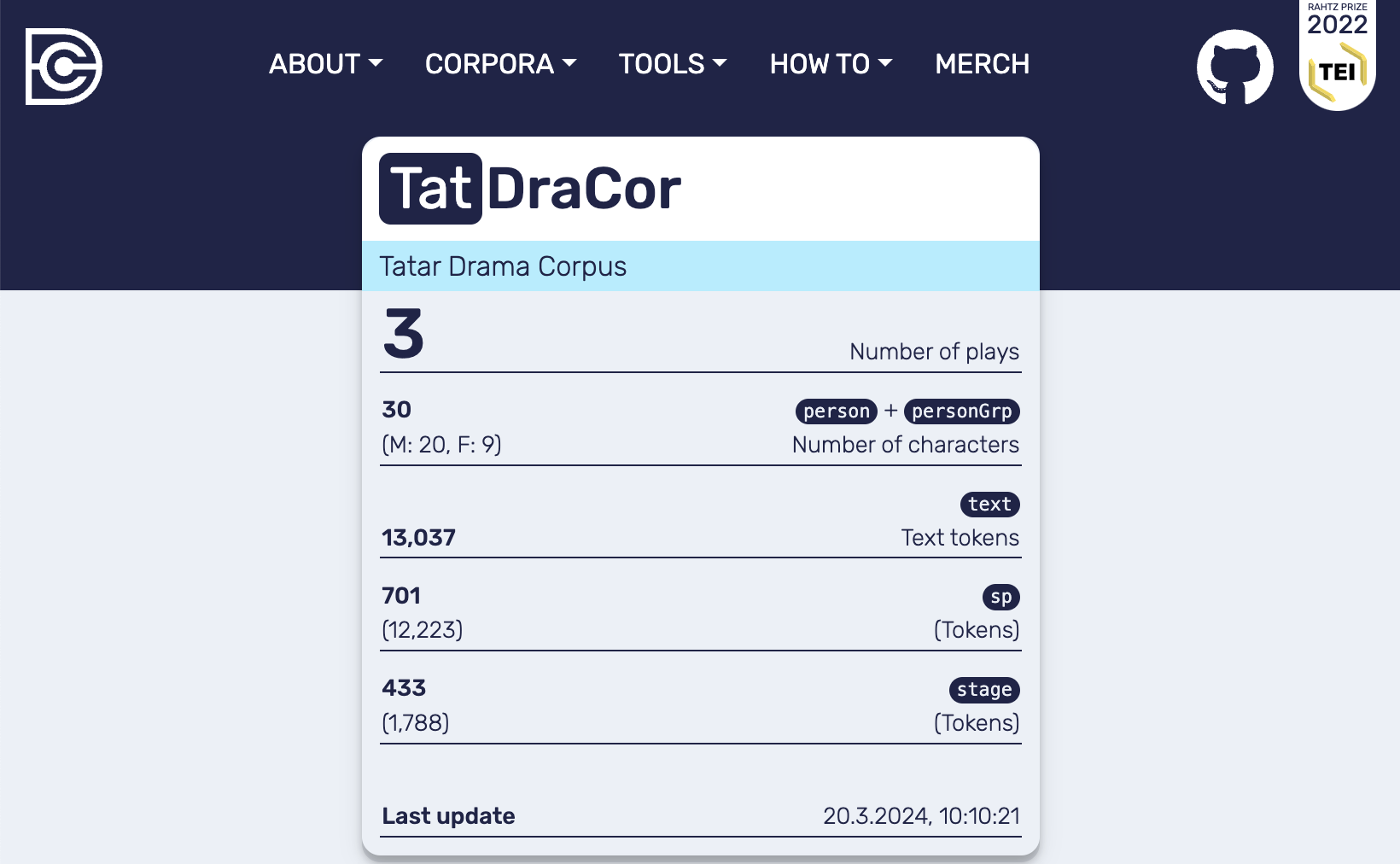
Fig. 11 Frontend of the local DraCor infrastructure with the “Tatar Drama Corpus” loaded#
It is also possible to directly add TEI files from the local filesystem, which allows a user to even use the DraCor environment with data not published on dracor.org or a public GitHub repository. When adding data to a local Docker container with the help of the “StableDraCor” package, the program keeps track of the constitution of the corpora and the sources used.
In the following code cell a single file is imported into the custom local corpus “FilesDraCor” (see Frontend of the local DraCor infrastructure with the additionally loaded file in the corpus “FilesDraCor”).
Show code cell content
# Create a corpus "FilesDraCor" and add a single play from the folder "import" to it
local_dracor.add_plays_from_directory(
corpusname="files",
directory="../import/"
)
!ls ../import
lessing-emilia-galotti.xml
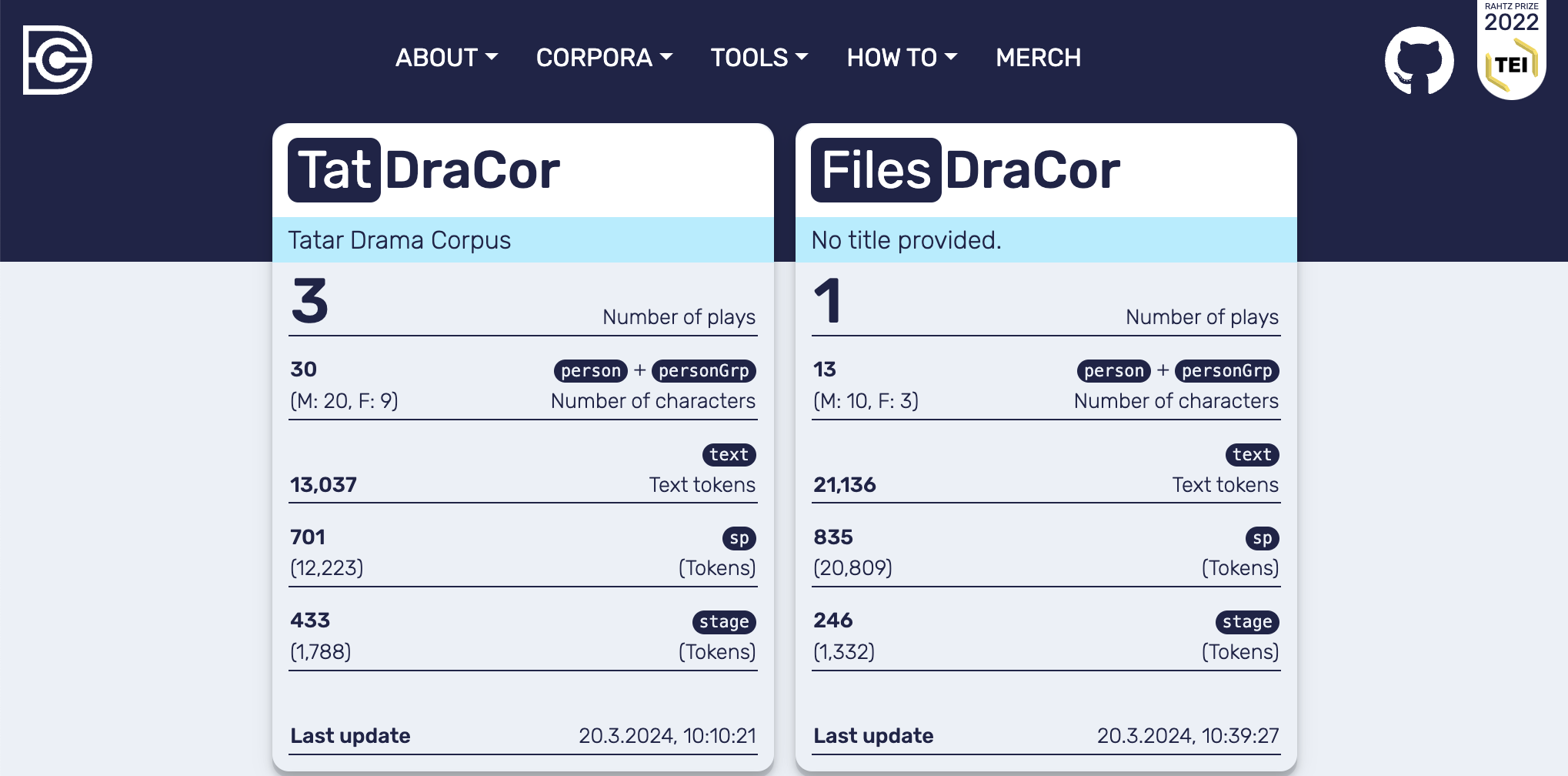
Fig. 12 Frontend of the local DraCor infrastructure with the additionally loaded file in the corpus “FilesDraCor”#
To allow for better reproducibility of the local infratructure it is recommended to used the functionality to to directly load corpora or parts thereof from a GitHub repository. This method of adding data allows to specify the “version” of the data in the corpus compilation process at a given point in time by referring to a single GitHub commit. As mentioned above, because DraCor corpora are “living corpora”, it is not guaranteed that corpora that are available on the web platform do not change. Therefore, it would not be a good idea to base research aiming at being repeatable at the data in the live system. By using data directly from GitHub with StableDraCor it is possible to include only the plays that were available, let’s say, two years ago and in the encoding state they were at this time.
In the following code cell the “Bashkir Drama Corpus” is added to the local database directly from its GitHub Repository.
Show code cell content
# Add the Bashkir Drama Corpus from GitHub in the version identified by a single commit
local_dracor.add_corpus_from_repo(
repository_name="bashdracor",
commit="c16b58ef3726a63c431bb9575b682c165c9c0cbd")
The local DraCor infrastructure that has been set up until this point can be explored at http://localhost:8088 (see Frontend of the local DraCor infrastructure with three corpora including the “Bashkir Drama Corpus”.
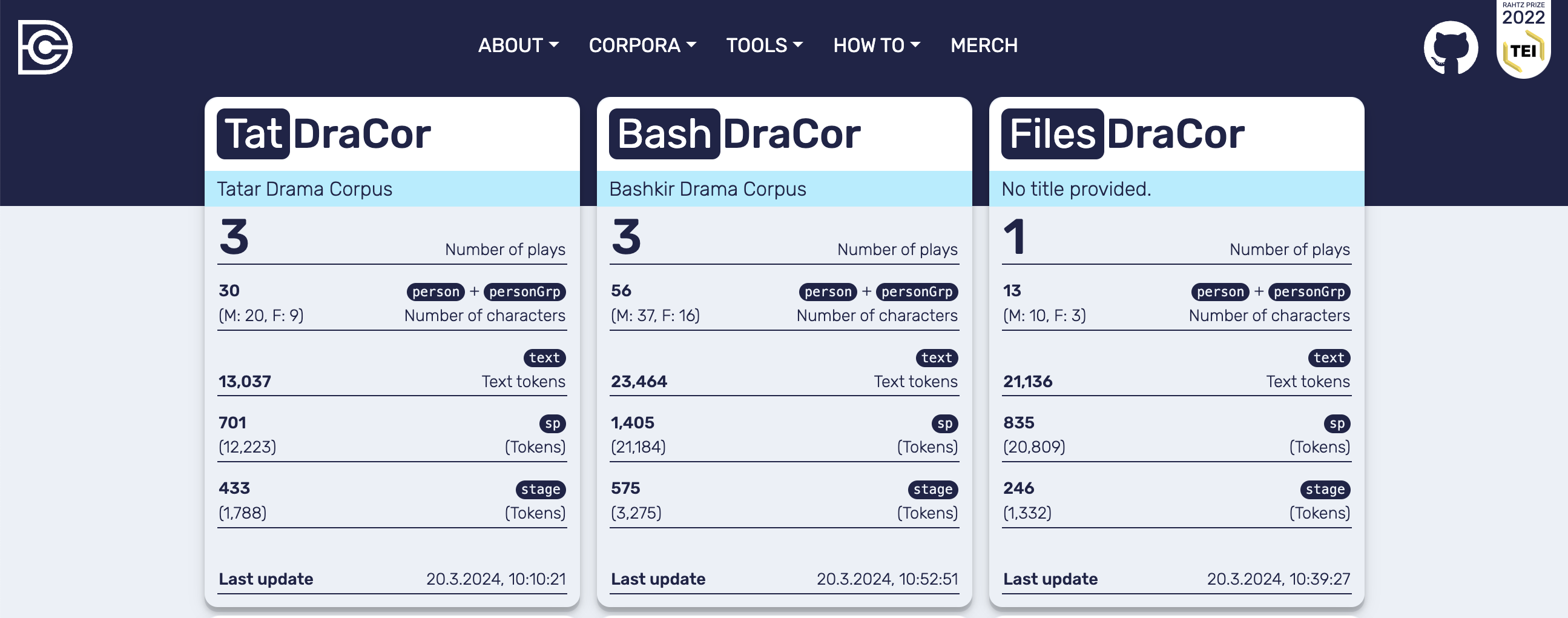
Fig. 13 Frontend of the local DraCor infrastructure with three corpora including the “Bashkir Drama Corpus”#
The tool keeps track of the whole configuration of the system: This includes the versions of the microservices used, and the corpora loaded. The state of the corpus is identified by a timestamp and – if the source is a GitHub repository, the commit. This “log” can be output as a “manifest” JSON object (command: get_manifest()), which should allow re-creating the system even if no Docker image is available. It would also allow a user to unambiguously identify the exact data that was used in a study.[13]
The following code snippet shows such a manifest[14] with several corpora added:
Show code cell source
# Output the manifest documenting the local DraCor System
local_dracor.get_manifest()
{'version': 'v1',
'system': {'id': 'da0980c8-0ed9-4120-9f72-6989906435b6',
'name': 'my_local_dracor',
'description': 'My local demo DraCor system',
'timestamp': '2024-04-29T07:24:12.185273'},
'services': {'api': {'container': '9508a2c75338',
'image': 'dracor/dracor-api:v0.90.1-local',
'version': '0.90.1-2-g19a3f46-dirty',
'existdb': '6.0.1'},
'frontend': {'container': '5cdeab5eb2c2',
'image': 'dracor/dracor-frontend:v1.6.0-dirty'},
'metrics': {'container': '3e39341abb86',
'image': 'dracor/dracor-metrics:v1.2.0'},
'triplestore': {'container': 'e054e879593f',
'image': 'dracor/dracor-fuseki:v1.0.0'}},
'corpora': {'tat': {'corpusname': 'tat',
'timestamp': '2024-04-29T07:23:57.313767',
'sources': {'tat': {'type': 'api',
'corpusname': 'tat',
'url': 'https://dracor.org/api/corpora/tat',
'timestamp': '2024-04-29T07:23:57.313775',
'num_of_plays': 3}},
'num_of_plays': 3},
'files': {'corpusname': 'files',
'timestamp': '2024-04-29T07:24:02.373231',
'sources': {'fd55d6f8': {'type': 'files',
'timestamp': '2024-04-29T07:24:02.373238'}},
'num_of_plays': 1},
'bash': {'corpusname': 'bash',
'timestamp': '2024-04-29T07:24:06.675530',
'sources': {'bash': {'type': 'repository',
'corpusname': 'bash',
'url': 'https://github.com/dracor-org/bashdracor',
'commit': 'c16b58ef3726a63c431bb9575b682c165c9c0cbd',
'timestamp': '2024-04-29T07:24:06.675539',
'num_of_plays': 3}},
'num_of_plays': 3}}}
“StableDraCor” supports creating a Docker image from a populated database container. With the original workflow it was necessary to do this with the docker commit command in the terminal. It was also necessary to provide additional documentation, for example the Jupyter notebook that was used to assemble a corpus. This information was ‘detached’ from the Docker image and it was necessary to explicitly point to this form of documentation, because it was not part of the research artifact itself. We tackled this issue with StableDracor and found a way to include machine readable documentation about the research artifact directly attached to it. Now, when we create an image with the tool, we issue a slightly different docker commit command that also attaches Docker Object Labels[15] directly to the newly created image. We achieve this by taking the manifest as mentioned earlier, and decomposing it into single Docker Labels. StableDraCor can convert a manifest into labels but also re-convert Docker Object Labels on the image in the org.dracor.stable-dracor.* namespace back into a manifest. By providing the manifest information as image labels, we allow a user, for example, to retrieve information about the corpus contents and the sources of a database without having to run the image as a Docker container first. We also attach the information about the individual DraCor microservices directly to the image as labels.
In the following code cell a Docker Image of the the DraCor API container is created.
Show code cell content
# Create a Docker image of the DraCor API container
local_dracor.create_docker_image_of_service(service="api",
image_tag="d73_demo")
WARNING:root:The dracor-api container is running. There might be issues with the image, if it is create from a running container. Consider stopping it before creating the image.
The labels attached to this image are as follows:
Show code cell outputs
{'com.docker.compose.config-hash': '2b3dcea466df52b1886c275c872b8b2aacf2b3646ddec2a6dee7a2bf2e6b6535',
'com.docker.compose.container-number': '1',
'com.docker.compose.depends_on': 'fuseki:service_started:false,metrics:service_started:false',
'com.docker.compose.image': 'sha256:171df59ae0ab650356c45feeabe2a65b63b77c3e9bd7bf362926bfdd78e931f8',
'com.docker.compose.oneoff': 'False',
'com.docker.compose.project': 'my_local_dracor',
'com.docker.compose.project.config_files': '-',
'com.docker.compose.project.working_dir': '/home/d73/report',
'com.docker.compose.service': 'api',
'com.docker.compose.version': '2.23.0',
'org.dracor.stable-dracor.corpora': 'tat,files,bash',
'org.dracor.stable-dracor.corpora.bash.corpusname': 'bash',
'org.dracor.stable-dracor.corpora.bash.num-of-plays': '3',
'org.dracor.stable-dracor.corpora.bash.sources': 'bash',
'org.dracor.stable-dracor.corpora.bash.sources.bash.commit': 'c16b58ef3726a63c431bb9575b682c165c9c0cbd',
'org.dracor.stable-dracor.corpora.bash.sources.bash.corpusname': 'bash',
'org.dracor.stable-dracor.corpora.bash.sources.bash.num-of-plays': '3',
'org.dracor.stable-dracor.corpora.bash.sources.bash.timestamp': '2024-04-29T07:24:06.675539',
'org.dracor.stable-dracor.corpora.bash.sources.bash.type': 'repository',
'org.dracor.stable-dracor.corpora.bash.sources.bash.url': 'https://github.com/dracor-org/bashdracor',
'org.dracor.stable-dracor.corpora.bash.timestamp': '2024-04-29T07:24:06.675530',
'org.dracor.stable-dracor.corpora.files.corpusname': 'files',
'org.dracor.stable-dracor.corpora.files.num-of-plays': '1',
'org.dracor.stable-dracor.corpora.files.sources': 'fd55d6f8',
'org.dracor.stable-dracor.corpora.files.sources.fd55d6f8.timestamp': '2024-04-29T07:24:02.373238',
'org.dracor.stable-dracor.corpora.files.sources.fd55d6f8.type': 'files',
'org.dracor.stable-dracor.corpora.files.timestamp': '2024-04-29T07:24:02.373231',
'org.dracor.stable-dracor.corpora.tat.corpusname': 'tat',
'org.dracor.stable-dracor.corpora.tat.num-of-plays': '3',
'org.dracor.stable-dracor.corpora.tat.sources': 'tat',
'org.dracor.stable-dracor.corpora.tat.sources.tat.corpusname': 'tat',
'org.dracor.stable-dracor.corpora.tat.sources.tat.num-of-plays': '3',
'org.dracor.stable-dracor.corpora.tat.sources.tat.timestamp': '2024-04-29T07:23:57.313775',
'org.dracor.stable-dracor.corpora.tat.sources.tat.type': 'api',
'org.dracor.stable-dracor.corpora.tat.sources.tat.url': 'https://dracor.org/api/corpora/tat',
'org.dracor.stable-dracor.corpora.tat.timestamp': '2024-04-29T07:23:57.313767',
'org.dracor.stable-dracor.services': 'api,frontend,metrics,triplestore',
'org.dracor.stable-dracor.services.api.base-image': 'dracor/dracor-api:v0.90.1-local',
'org.dracor.stable-dracor.services.api.existdb': '6.0.1',
'org.dracor.stable-dracor.services.api.image': 'dracor/stable-dracor:d73_demo',
'org.dracor.stable-dracor.services.api.version': '0.90.1-2-g19a3f46-dirty',
'org.dracor.stable-dracor.services.frontend.image': 'dracor/dracor-frontend:v1.6.0-dirty',
'org.dracor.stable-dracor.services.metrics.image': 'dracor/dracor-metrics:v1.2.0',
'org.dracor.stable-dracor.services.triplestore.image': 'dracor/dracor-fuseki:v1.0.0',
'org.dracor.stable-dracor.system.description': 'My local demo DraCor system',
'org.dracor.stable-dracor.system.id': 'da0980c8-0ed9-4120-9f72-6989906435b6',
'org.dracor.stable-dracor.system.name': 'my_local_dracor',
'org.dracor.stable-dracor.system.timestamp': '2024-04-29T07:24:12.226746',
'org.dracor.stable-dracor.version': 'v1'}
In summary, our “StableDraCor” package allows to generate a fully self-describing, completely versionized research artifact that alone is sufficient to replicate the corpora and their research infrastructure that were used in a study.
4.4 Practical Examples#
The following section demonstrates how the concepts and tools introduced in this report can be used to facilitate future repeating of research based on DraCor infrastructure components and corpora. The first example shows how to identify and then stabilize the exact version of the corpus used in a research paper that was discussed earlier in section 3.1 How to Better Not Cite a Living Corpus. An Example From Current Research. In the second example a version of the “DLINA Corpus Sydney” is created on the basis of recent DraCor data to allow for repeating research carried out in the context of the DLINA project on the same set of plays.[16]
4.4.1 Reconstructing (and Stabilizing) Corpora Used to Train and Evaluate a Classifier for Chiasmus Detection#
In 3.1 How to Better Not Cite a Living Corpus. An Example From Current Research the paper „Data-Driven Detection of General Chiasmi Using Lexical and Semantic Features“ [Schneider et al., 2021] is discussed as an example of research that re-uses the German Drama Corpus. The authors do not use the DraCor API for their study but download data directly from the GerDraCor GitHub Repository. The only information that hints at which version of the corpus was used to train and test the classifier is the number of plays that were included in the corpus at the time. The authors report that GerDraCor included 504 plays.
In the following code cells we use the “corpus archeology script” described in the section Excursus: An Algorithmic Archaeology of a Living Corpus: GerDraCor as a Dynamic Epistemic Object to identify the actual version of the German Drama Corpus used and load this version into a local DraCor infrastructure as described in section 4.3 Simplifying the Workflow: StableDraCor:
Show code cell content
# Start the analysis with previously downloaded data
repo = GitHubRepo(repository_name="gerdracor",
github_access_token=github_token,
import_commit_list="tmp/gerdracor_commits.json",
import_commit_details="tmp/gerdracor_commits_detailed.json",
import_data_folder_objects="tmp/gerdracor_data_folder_objects.json",
import_corpus_versions="tmp/gerdracor_corpus_versions.json")
# To get the version with 504 plays:
# Get the versions as a dataframe containing the number of plays included ("document_count")
play_counts_df = repo.get_corpus_versions_as_df(columns=["id","date_from","document_count"])
# Filter the dataframe on versions that have exactly 504 plays
play_counts_df[play_counts_df["document_count"] == 504]
| id | date_from | document_count | |
|---|---|---|---|
| 1212 | 6e1020dcfcb98a0d027ceb401a6a5fbd4537fe29 | 2020-09-26 11:47:04+00:00 | 504 |
The most probable version of the GerDraCor data used is identified by the SHA value 6e1020dcfcb98a0d027ceb401a6a5fbd4537fe29 and dates from 26 September, 2020.[17] In the following cell a Docker container of the DraCor API is created and populated with this exact corpus version (see Frontend of the local DraCor infrastructure with GerDraCor (version: 6e1020dcfcb98a0d027ceb401a6a5fbd4537fe29) including 504 plays):
Show code cell content
# Initialize the StableDraCor instance; add metadata
chiasmus_detection_dracor = StableDraCor(
name="chiasmus_detection",
description="DraCor system including the GerDraCor version used in the paper 'Data-Driven Detection of General Chiasmi'",
github_access_token=github_token
)
# Run the infrastructure
chiasmus_detection_dracor.run()
# Add the corpus version
# Because data is ingested (which is a slow process) this will take some time (approx. 30min!)
# Be very patient. You can check http://localhost:8088 to see the progress. The number of plays
# in the corpus should constantly increase when reloading the page.
chiasmus_version_commit_id = play_counts_df[play_counts_df["document_count"] == 504].iloc[0]["id"]
chiasmus_detection_dracor.add_corpus_from_repo(
repository_name="gerdracor",
commit=chiasmus_version_commit_id)
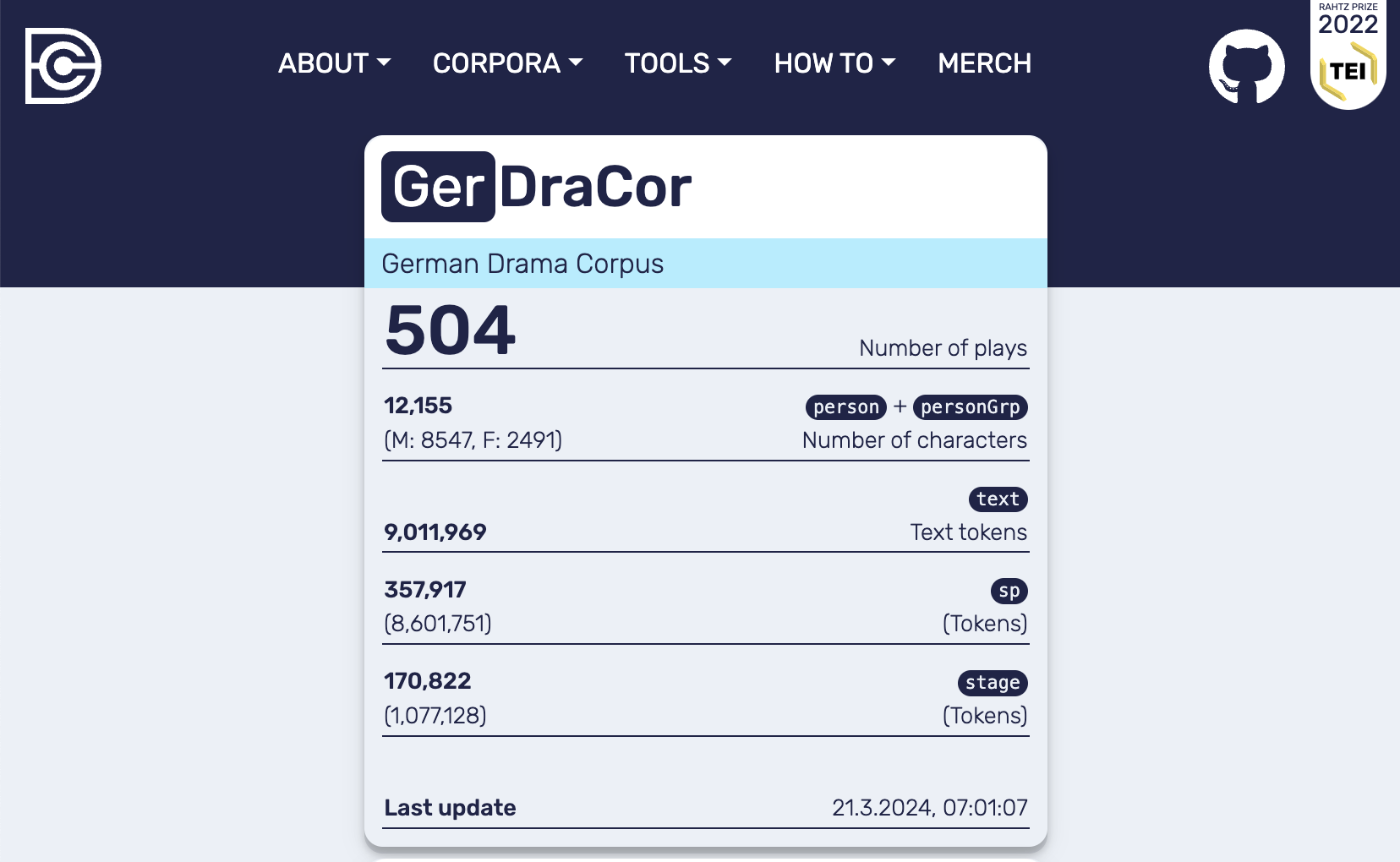
Fig. 14 Frontend of the local DraCor infrastructure with GerDraCor (version: 6e1020dcfcb98a0d027ceb401a6a5fbd4537fe29) including 504 plays#
For training of the classifier a manually annotated data set consisting of four plays by the author Friedrich Schiller are used (cf. [Schneider et al., 2021, p.98]; citations can also be found in 3.1 How to Better Not Cite a Living Corpus. An Example From Current Research) In the paper the titles of these plays are included. In the following list DraCor identifiers are added:
Die Piccolomini (
schiller-die-piccolomini, ger000086)Wallensteins Lager (
schiller-wallensteins-lager, ger000025)Wallensteins Tod (
schiller-wallensteins-tod, ger000058)Wilhelm Tell (
schiller-wilhelm-tell, ger000452)
Show code cell content
# [...] four annotated texts by Friedrich Schiller
# Die Piccolomini,
# Wallensteins Lager,
# Wallensteins Tod
# and Wilhelm Tell.
# Add an empty new corpus "training" with the following metadata
chiasmus_annotated_corpus_metadata = {
"name" : "training",
"title": "Schiller Training Corpus",
"description": "Corpus of four plays by Friedrich Schiller used to train the Chiasmus Classifier"
}
chiasmus_detection_dracor.add_corpus(corpus_metadata=chiasmus_annotated_corpus_metadata)
# Create a list with the playnames/filenames of the plays to add
chiasmus_annotated_schiller_corpus_playnames = [
"schiller-die-piccolomini",
"schiller-wallensteins-lager",
"schiller-wallensteins-tod",
"schiller-wilhelm-tell"]
# Add each play in the respective version to the previously created corpus
for playname in chiasmus_annotated_schiller_corpus_playnames:
chiasmus_detection_dracor.add_play_version_to_corpus(
filename=playname,
repository_name="gerdracor",
commit=chiasmus_version_commit_id,
corpusname="training")
The classifier is then tested on a data set consisting of the other seven plays by Friedrich Schiller included in the GerDraCor corpus at that time. The authors do not explicitly spell out the titles but with having the local instance of GerDraCor one can filter for the plays by Schiller or even use the API to filter out plays by a certain author (identified by a Wikidata Identfier, here: Q22670) as is demonstrated in the following hidden code cell. We use this information to create an additional corpus test in the local DraCor instance.
Show code cell content
# [...] seven other dramas by Friedrich Schiller [...]
# Add an empty new corpus "test" with the following metadata
chiasmus_test_corpus_metadata = {
"name" : "test",
"title": "Schiller Test Corpus",
"description": "Corpus of the seven other plays by Friedrich Schiller used to test the Chiasmus Classifier"
}
chiasmus_detection_dracor.add_corpus(corpus_metadata=chiasmus_test_corpus_metadata)
# Create a list with the playnames/filenames of the plays to add
chiasmus_schiller_testset_playnames = [
"schiller-maria-stuart",
"schiller-kabale-und-liebe",
"schiller-don-carlos-infant-von-spanien",
"schiller-die-verschwoerung-des-fiesco-zu-genua",
"schiller-die-raeuber",
"schiller-die-jungfrau-von-orleans",
"schiller-die-braut-von-messina"]
# Add each play in the respective version to the previously created corpus
for playname in chiasmus_schiller_testset_playnames:
chiasmus_detection_dracor.add_play_version_to_corpus(
filename=playname,
repository_name="gerdracor",
commit=chiasmus_version_commit_id,
corpusname="test")
The last corpus that is added to the local DraCor instance contains the remaining 493 documents from GerDraCor that are also used in the paper for testing purposes. In the hidden code cell below this corpus called rest is created:
Show code cell content
# "[...] remaining 493 documents from GerDraCor" therefore nedd to exclude
# all the plays by Friedrich Schiller in the two lists above
chiasmus_playnames_exclude = chiasmus_annotated_schiller_corpus_playnames + chiasmus_schiller_testset_playnames
chiasmus_rest_corpus_metadata = {
"name" : "rest",
"title": "Non-Schiller plays",
"description": "Corpus of the remaining plays in GerDraCor used to test the Chiasmus Classifier"
}
# Add the GerDraCor version, but change the Corpus Metadata; exclude all Schiller-plays added before as separate corpora
chiasmus_detection_dracor.add_corpus_from_repo(
commit = chiasmus_version_commit_id,
repository_name = "gerdracor",
use_metadata_of_corpus_xml = False,
corpus_metadata = chiasmus_rest_corpus_metadata,
exclude = chiasmus_playnames_exclude)
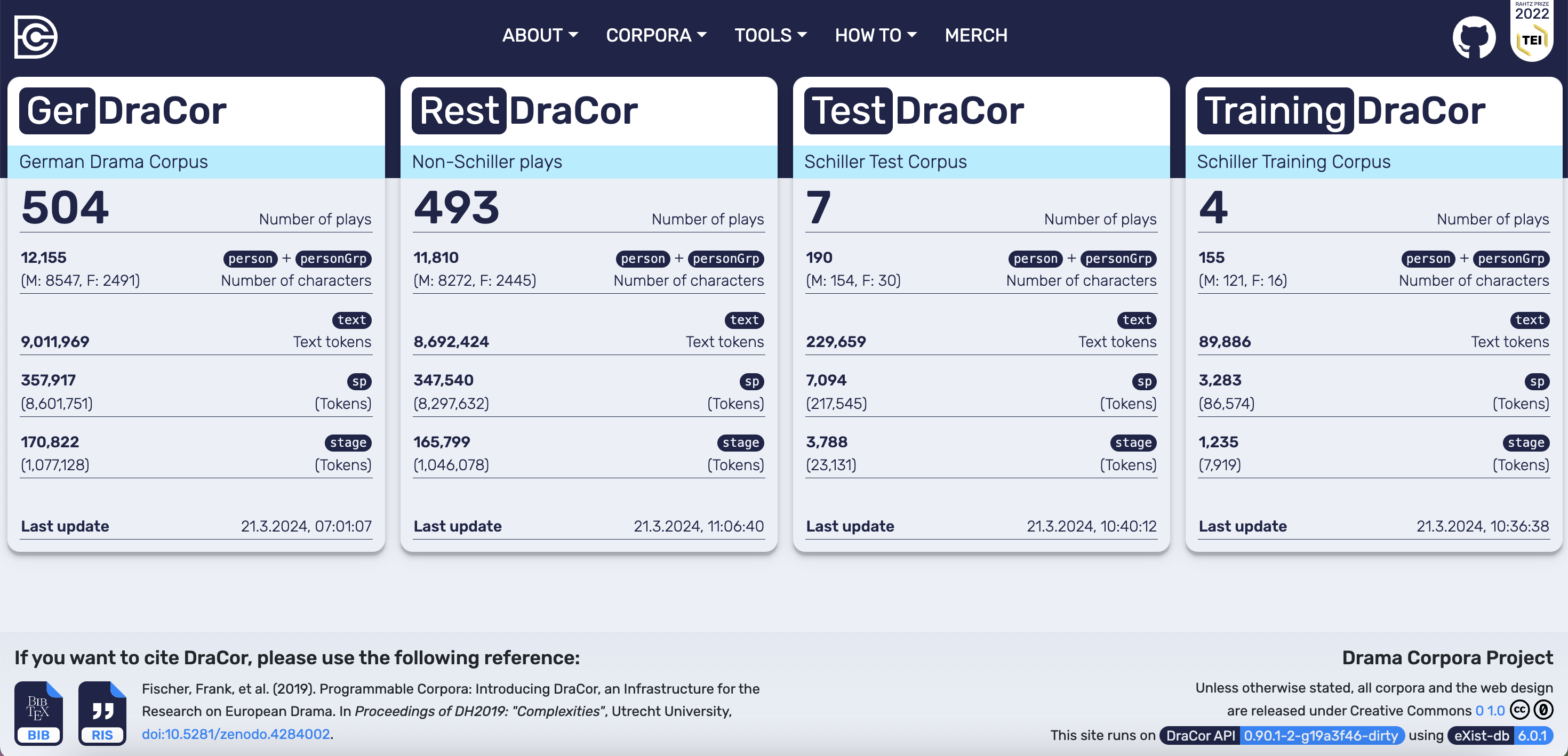
Fig. 15 Frontend of the local DraCor infrastructure including the corpora mentioned in the “Chiasmus Detection” paper#
4.4.2 GerDraCor-based DLINA Sydney Corpus#
In section 3.1 The “Birth” of GerDraCor the DLINA Sidney Corpus was mentioned as the main source of the later GerDraCor corpus. The original DLINA corpus contained 465 plays taken from the TextGrid Repository. As the corpus archeology (see Batch Edits as Major Revisions) has shown there were several significant changes to the encoding which would result in the early added data not being compatible with the current versions of the DraCor API. Still, it might be an interesting use case to repeat earlier studies of the DLINA collective on the basis of a recent version of the data. Therefore, the following code samples demonstrate how a GerDraCor-based DLINA corpus can be built in a local DraCor setup. It will include all 465 plays originally included in the DLINA corpus, but in the versions as are included in the latest GerDraCor.
Show code cell content
dlina_gerdracor = StableDraCor(
name="dlina_gerdracor",
description="DraCor system including all plays available in the DLINA Sydney corpus in a recent DraCor encoding",
github_access_token=github_token
)
dlina_gerdracor.run()
With the commit “fdac66ba90c2c094012dc90395e952411d324e4c” the original DLINA file names of all TEI-XML files are changed to now match the identifier playname in GerDraCor, but after that, there are still some files renamed.
# Should get the list of the original playnames,
# but make sure there are no problems with renamed plays
version_renaming_id = "fdac66ba90c2c094012dc90395e952411d324e4c"
# From the dictonary representing the "version" get the field "playnames" that contains the
# plays available in a certain version
dlina_playnames = repo.get_corpus_version(version=version_renaming_id)["playnames"]
# Get the file-names from the renaming version
rename_incidents = repo.get_renamed_files(
exclude_versions=["e18c322706417825229f1471b15bd6daaeaf3ab1",
"fdac66ba90c2c094012dc90395e952411d324e4c"])
new_gerdracor_playnames = []
unchanged_playnames = []
for original_playname in dlina_playnames:
renamed_flag = False
for item in rename_incidents:
if f"tei/{original_playname}.xml" == item["previous_filename"]:
new_gerdracor_playname = item["new_filename"].split("/")[1].replace(".xml","")
new_gerdracor_playnames.append(new_gerdracor_playname)
renamed_flag = True
if renamed_flag is False:
unchanged_playnames.append(original_playname)
# Get the latest commit to the GerDraCor repository.
# This will be included in the description of the new corpus
# it is also needed because this version will be used as source of the plays added to the corpus
latest_gerdracor_version_id = repo.get_latest_corpus_version()["id"]
# Add an empty new corpus "dlina" with the following metadata
dlina_corpus_metadata = {
"name" : "dlina",
"title": "GerDraCor-based DLINA Corpus Sydney",
"description": f"Version of the German Drama Corpus (GerDraCor, version {latest_gerdracor_version_id}) containing only plays available in the DLINA Corpus Sydney"
}
dlina_gerdracor.add_corpus(corpus_metadata=dlina_corpus_metadata)
True
# Add each play in the respective version to the previously created corpus
playnames_to_add = new_gerdracor_playnames + unchanged_playnames
for playname in playnames_to_add:
dlina_gerdracor.add_play_version_to_corpus(
filename=playname,
repository_name="gerdracor",
commit=latest_gerdracor_version_id,
corpusname="dlina")
WARNING:root:Could not add play from source 'https://raw.githubusercontent.com/dracor-org/gerdracor/2def59bb9123187740bf4c3136eb500deee154db/tei/fouque-der-held-des-nordens.xml'.
# This is not a renaming problem with fouque-der-held-des-nordens;
# the play has been split up in three
# Mir fällt schon auf, Ich frag hier immer wegen DLINA... also https://dlina.github.io/linas/212/ gibt es so nicht mehr; das ist kein Fall von "Umbenennung"; ist das aus dem ursprünglichen lina212 ->
# TEI version ger000212, ger000471 und ger000470 geworden?
#pt jip, das ist in DraCor gesplittet
# ff: a, die 5 mehrteiligen stücke in dieser liste (von 2015) hatten wir schon vorher gesplittet:https://github.com/DLiNa/project/blob/master/data/TextGrid-Repository---List-of-all-dramatic-texts.txt
# (in der liste die nummern 18, 137, 217, 273, 490 – das waren aber noch keine dracor-IDs)
#bei fouqués held des nordens fiel mir die mehrteiligkeit dann erst auf, als ich stück für stück durchgegangen bin, daher das spätere splitting…
# Sigurd, der Schlangentödter
# "fouque-sigurds-rache"
# Aslauga
# The DLINA play https://dlina.github.io/linas/212/ is actually a trilogy;
#In GerDraCor the play is split into three indiviual plays
fouque_held_des_nordens_part_playnames = [
"fouque-sigurd-der-schlangentoedter",
"fouque-sigurds-rache",
"fouque-aslauga"
]
for playname in fouque_held_des_nordens_part_playnames:
dlina_gerdracor.add_play_version_to_corpus(
filename=playname,
repository_name="gerdracor",
commit=latest_gerdracor_version_id,
corpusname="dlina")
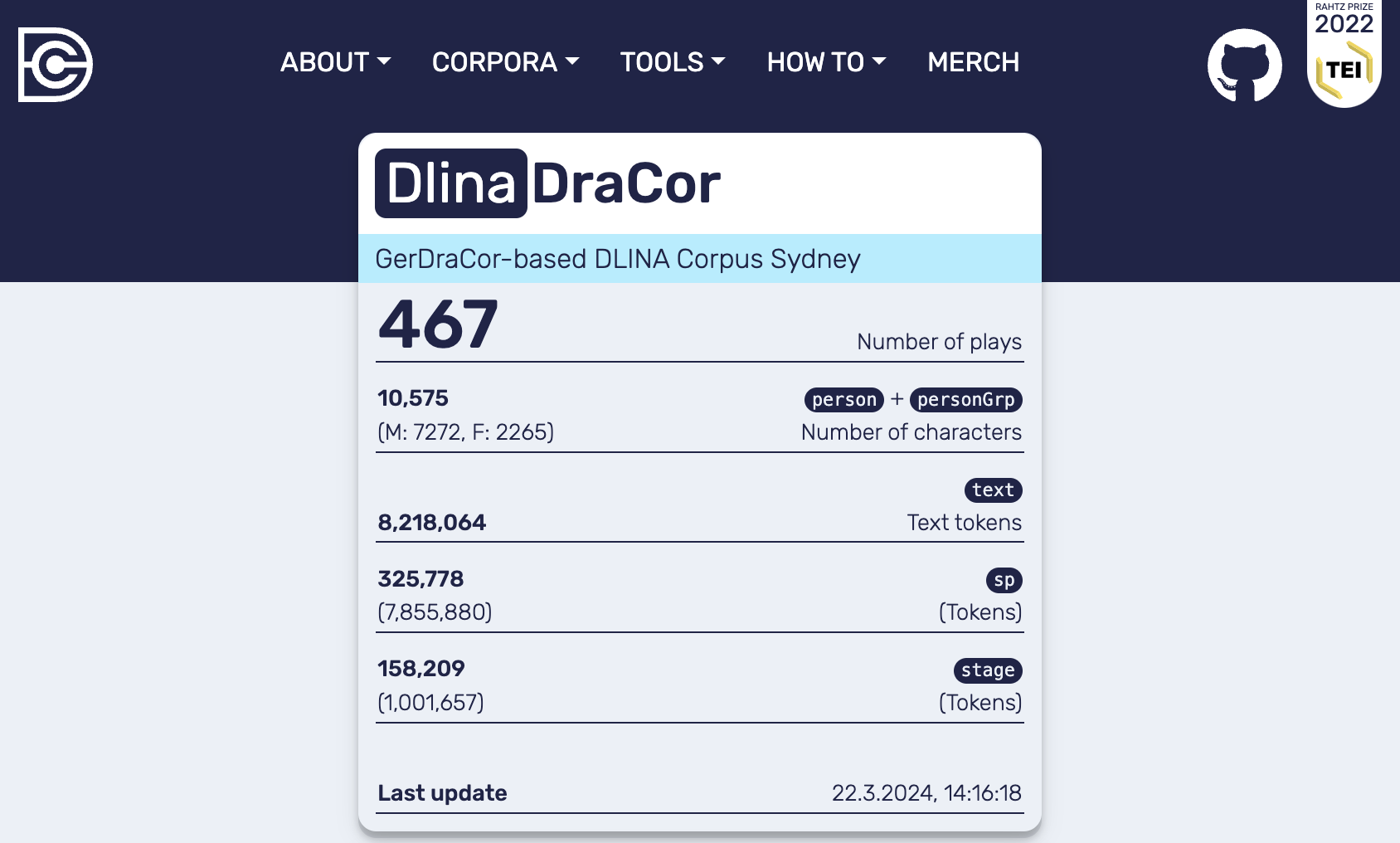
Fig. 16 Frontend of the local DraCor infrastructure including the plays contained in the DLINA Corpus Sydney#
# Get links to the commits that add the three individual plays
for playname in fouque_held_des_nordens_part_playnames:
print(repo.get_github_commit_url_of_version(
repo.get_corpus_version_adding_play(playname=playname)["id"]))
On December 16th, 2018 the play with the filename fouque-der-held-des-nordens.xml which is actually a trilogy was split up into three individual plays.
%%bash
# Stop and remove all Docker containers to avoid conflicts
# especially regarding ports in the next section
# This cell does not show up in the final rendering of the report
# If you want to use the Docker containers above and play around with them,
# the following commands should NOT be run
docker stop $(docker ps -a -q)
docker rm $(docker ps -a -q)
